BioinformaticS Software engineer
Hi! I’m Karen — a software engineer and ML enthusiast who enjoys building reliable systems people actually want to use.
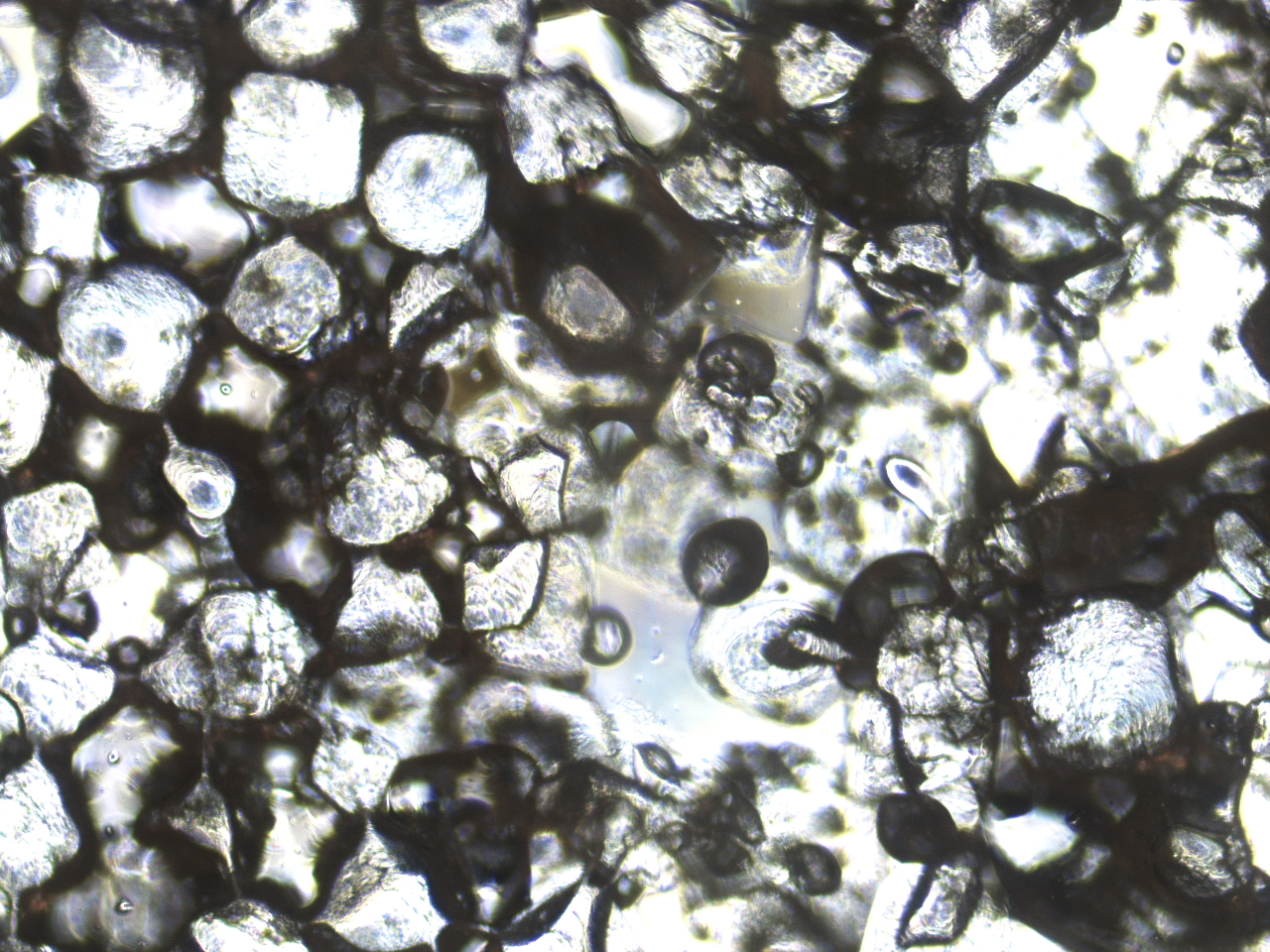
PLLA scaffolds for 3D Bone Tissue Engineering
I’m a software engineer who enjoys building tools that make scientific and data-heavy workflows feel simple! At the University of Washington I attained a B.S. in Bioengineering + Data Science. I am currently pursuing a Masters in Computer Science with a specialty in Machine Learning at Georgia Tech, expected August 2026!
I have over two years of industry experience in full-stack applications, automation pipelines, and ML-powered analysis tools for vaccinology and RNA research.
Computational projects
Doggy Database Full Stack Web App

Doggy Database is a full stack web application designed to manage dog adoptions, shelter operations, and reporting for an animal shelter. The system is built using a modern microservices architecture, leveraging Docker for containerization, PostgreSQL for data storage, Flask for backend APIs, and React for the frontend user interface.
Deep Learning for RNA Secondary Structure Prediction
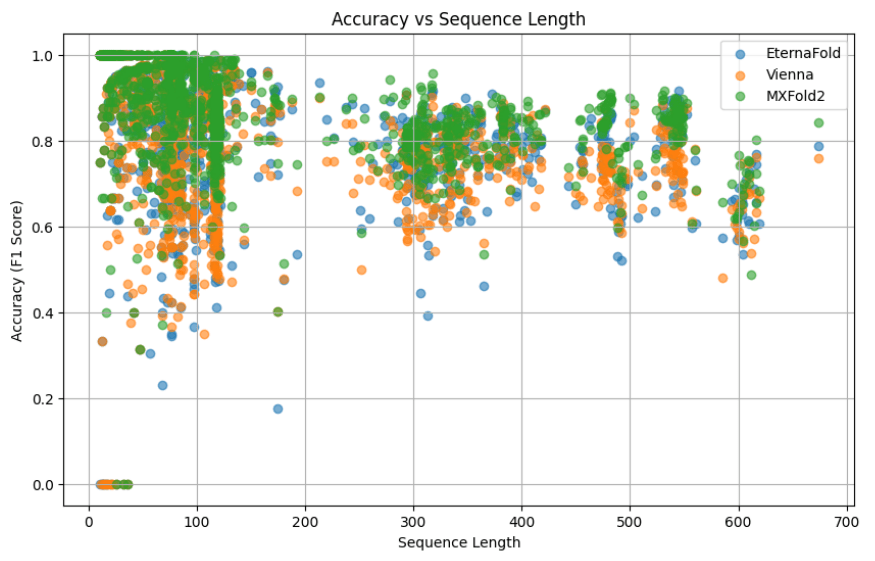
Analyzed deep learning models (EternaFold, MXFold2) for RNA structure prediction across varying sequence lengths and motifs. Utilized Mxfold2 to identify new sequence liabilities for RNA constructs.
Improved model robustness through hyperparameter tuning and data augmentation. Implemented super ensemble methods combining predictions from multiple models using weighted voting.
Github: Deep Learning for RNA Secondary Structure Prediction
A COVID-19 Vaccination Database using Microsoft Azure Portal
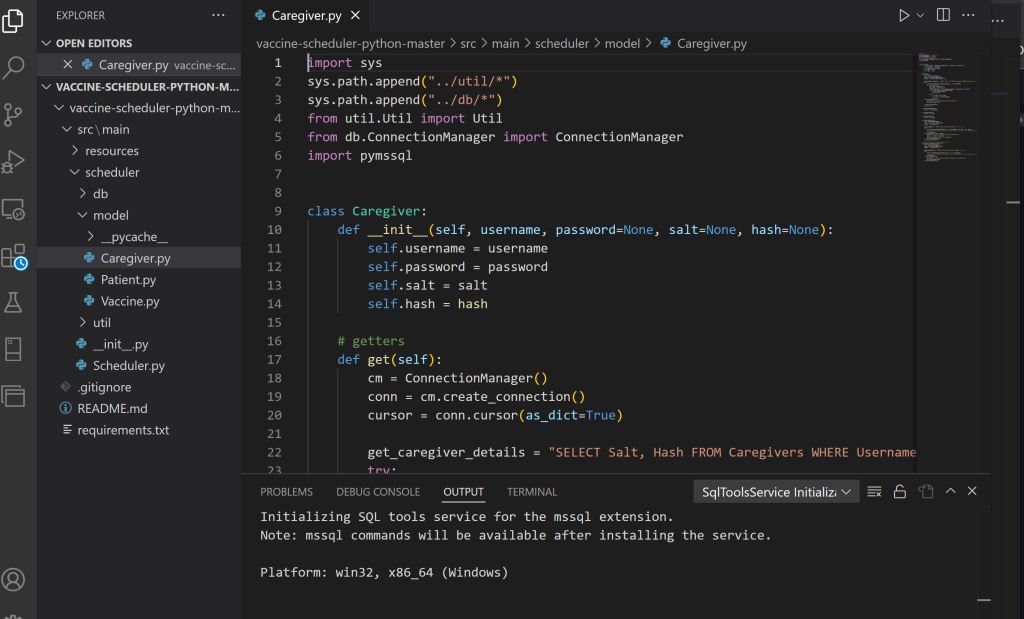
Developed an appointment reservation application that runs on the command line terminal and connects to a Microsoft Azure database server to help caregivers organize vaccinations. Wrote SQL commands and Python code within VS Code to update a server database via pymssql. Kept track of inventory, usernames, logins/logouts, appointments, and securely stored passwords by salting and hashing. Coded relational databases, transactions & commits, two-phase locking, and key management.
Machine Learning Methods and Manual Classification to Estimate Protein Secondary Structure

The protein function of a significant part of sequenced genomes are unknown. This is due to the high variability in secondary structures and functions that can result from the same subsequence. Consequently, a computational method for estimating secondary based on sequence provides a potent tool in resolving the structure and function of yet uncharacterized protein sequences. Primary protein sequence data was curated from the Protein Data Bank and mapped to secondary structure data generated by the DSSP. Sklearn was utilized to generate a decision tree classifier model on encoded categorical data compiled and cleaned from Kaggle. The model was tuned with hyperparameters on frame size and tree depth. Statistical analysis was performed on error and data was visualized with seaborn.
GitHub: Machine Learning Methods to Predict Protein Secondary Structure
wet lab research
Investigating the Effects of Mechanical Loading During Development in 3D Tissue-Engineered Bone

We report a novel, three-dimensional bone tissue model as a platform for musculoskeletal disease modeling that allows for compressive loading. By seeding induced pluripotent stem cell (iPSC) derived osteoblasts in a 3D, plasma treated, porous, gelatin coated, poly-L-lactide scaffold, we generated a 3D bone tissue culture with extracellular calcium deposition. By applying cyclical compression with an Arduino actuation device, we observed an improved bone phenotype. This method of generating 3D bone tissue may serve as a fast and accessible model for the formation of both healthy and pathological bone.
David Mack Lab at the Institute for Stem Cell and Regenerative Medicine
A Rainbow Lineage Reporter to Track Human Induced Pluripotent Stem Cells
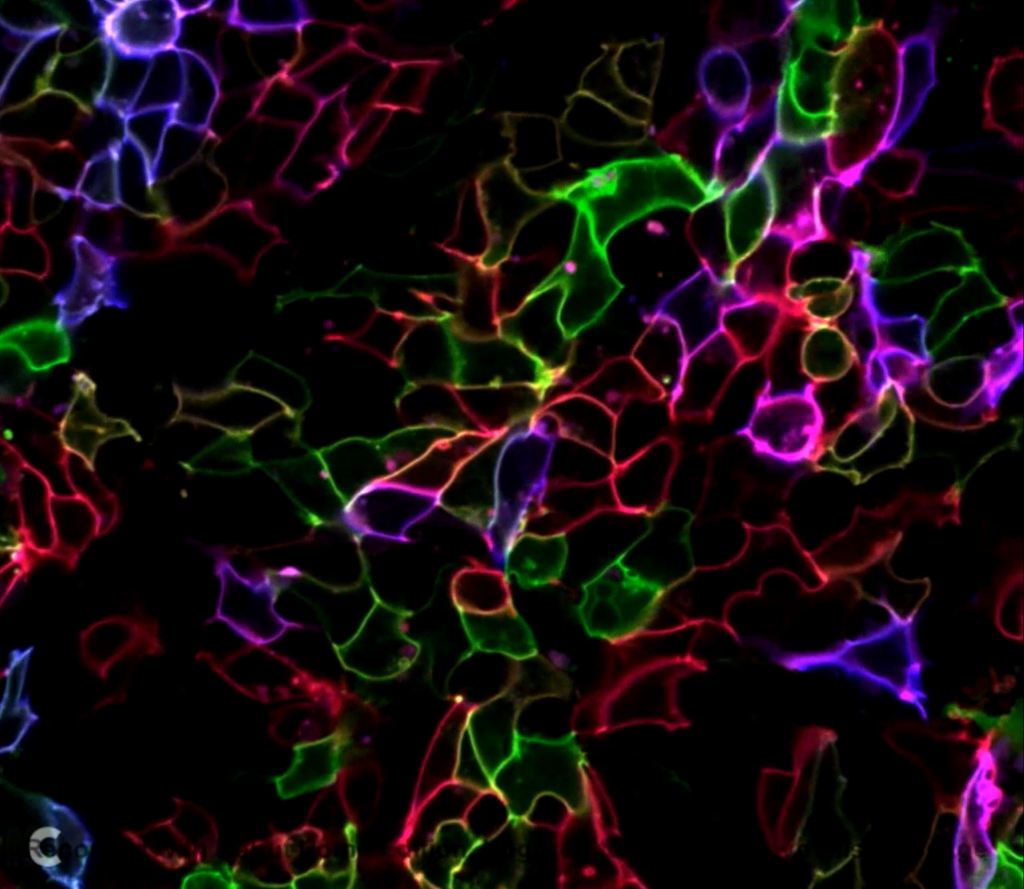
Identifying subsets of phenotypes in iPSCs and their differentiated progeny allow us to optimize tissue models for research. Here, we generated a rainbow reporter line in iPSCs that can track individual cells as they clonally expand and differentiate while providing phenotypic information. Knocking in four copies of a cassette containing three distinct fluorescent proteins allowed the expression of up to eighteen different colors. To date, we were able to create eight color barcoded iPSC lines for further experimentation. Color coded cells will allow us to track how the initial location/physical stresses/phenotype of an iPSC-derived cardiac cell in an engineered tissue determines its tissue layer and cell type.
Cell Biomechanics Lab at the Institute for Stem Cell and Regenerative Medicine
Publications and Awards
“Clade 2.3.4.4b but not historical clade 1 HA replicating RNA vaccine protects against bovine H5N1 challenge“
“A gH/gL-encoding replicon vaccine elicits neutralizing antibodies that protect humanized mice against EBV challenge “
“A Change of Heart: Human Cardiac Tissue Engineering as a Platform for Drug Development”
University of Washington Bioengineering Departmental Honors 2022
Mary Gates Research Scholar 2020
Mary Gates Research Scholar 2019
→ Bioexplore Mentor Program
Mentored UW bioengineering sophomores. Met on a bi-weekly basis to prepare for lab/internship positions and find their niche in the vast field of bioengineering.
→ Bioengage Outreach Facilitator
Traveled to Seattle World School, a school for low-income immigrants and refugees, to facilitate modules on bioengineering concepts on a bi-monthly basis.
Helped create lesson plans for 14-15 year olds where research-level engineering concepts were refined to interactive and educational modules.
→ Teaching Assistant BIOEN 326: Solid & Gel Mechanics
Taught a section of 17 sophomore bioengineering students for 1.5 hours a week. Facilitated study halls, held individual office hours, graded homework, exams, provided academic and career support.

Community Work
Expanding STEM education to under-represented communities and engaging with young scientists